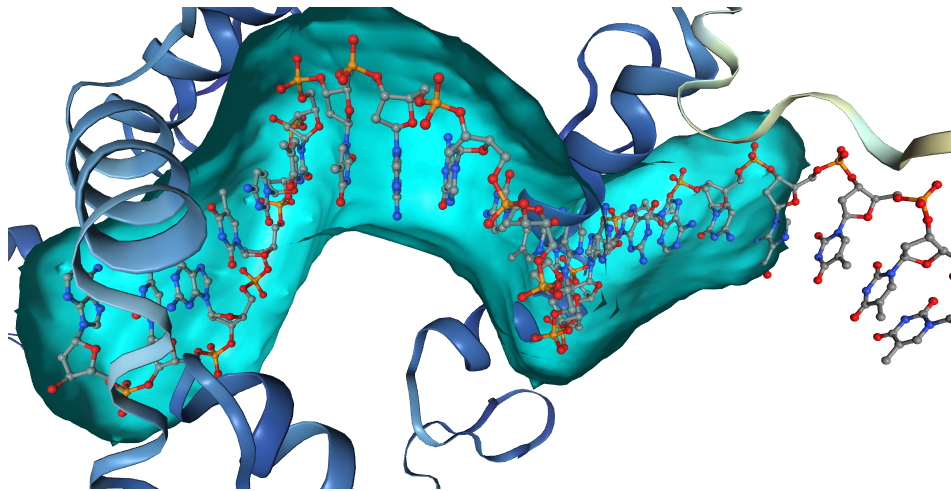
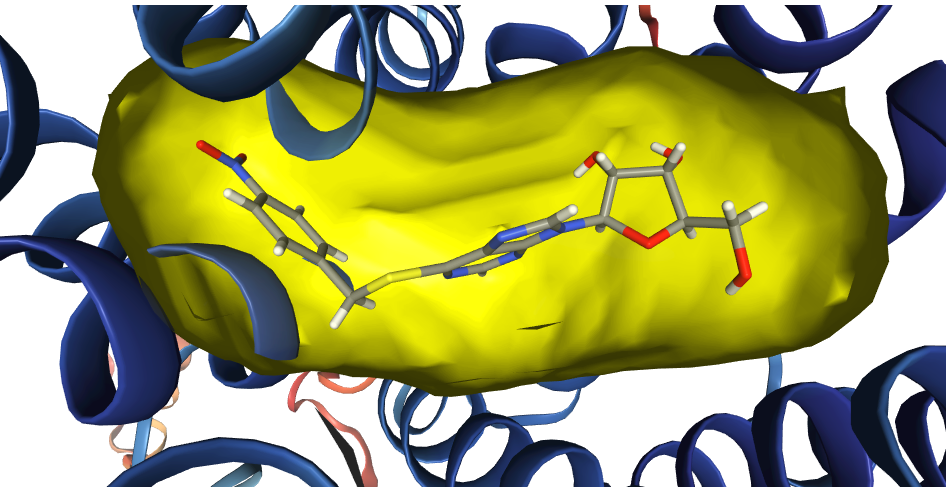
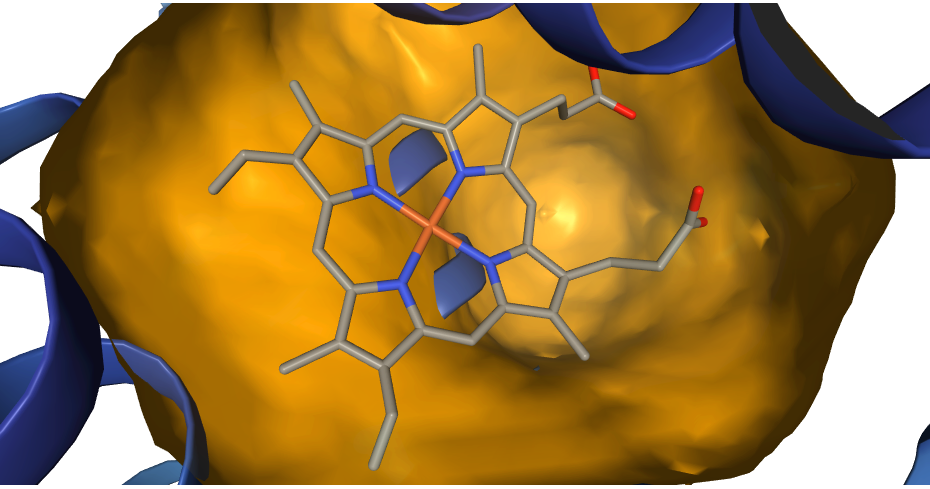
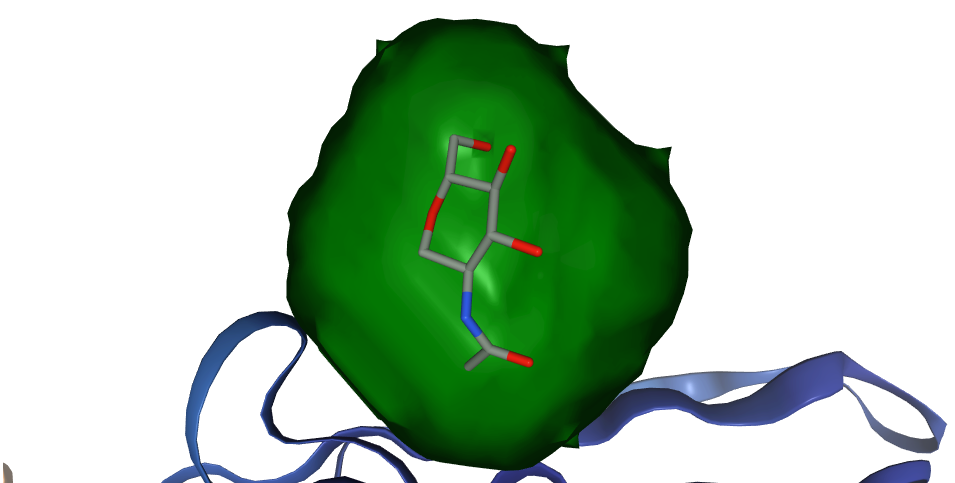
ProBiS-Fold annotates AlphaFold human protein database with
- Binding sites for: compounds (small molecules), cofactors, proteins, peptides, nucleic acids, metal ion and conserved water
- Post-translational modification sites (glycosylation sites)
- Predicted ligands and glycosides for each binding site (3D structures as bound to protein)
ProBiS-Fold aims to
- Provide interactive, downloadable binding sites for human proteome for functional and drug discovery studies
- Enable human proteome-wide structure-based virtual screening and selectivity prediction
Binding sites and post-translational sites types
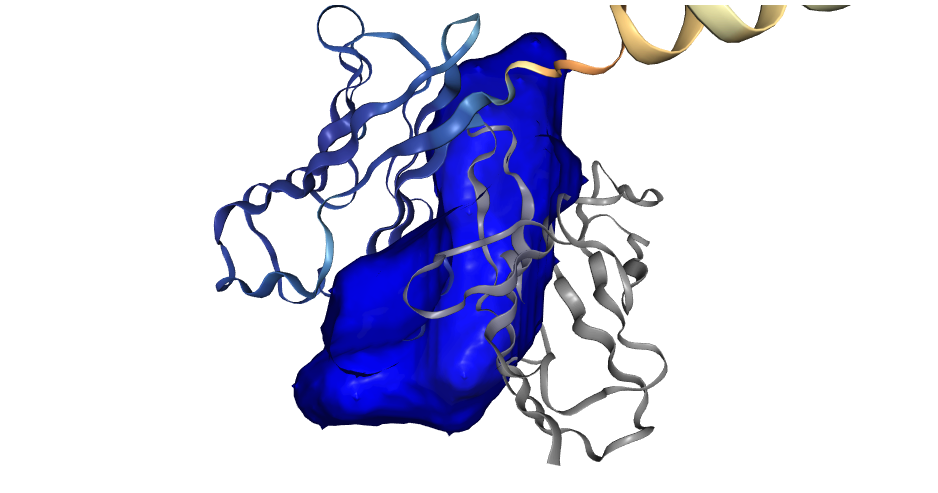
- Compound (substrate/agonist-competitive ligands), cofactor (cofactor and cofactor-competitive ligands) (based on list of known cofactors), protein (both <20 aa. and >=20 aa.), peptide (<20 aa.), nucleic acid (DNA or RNA molecules), metal ion (structurally conserved) and water (structurally conserved)
- Glycoslyation sites (O- and N- glycosylation)
- Ranked according to the estimated druggability score (applies to compound and cofactor sites)
Output
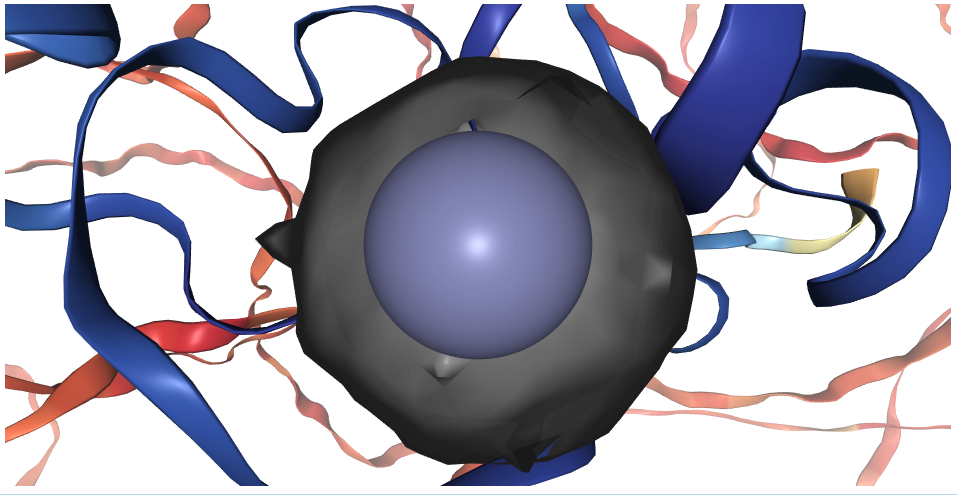
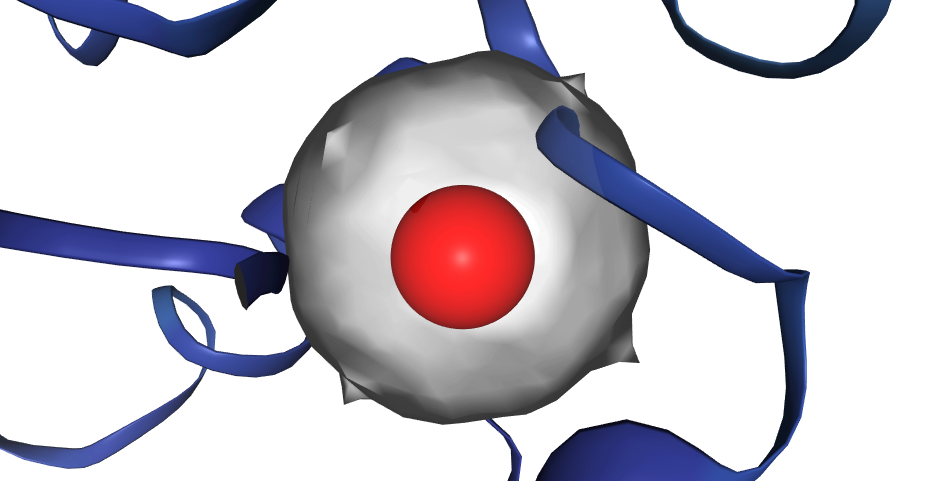
- Centroids (x,y,z,radius) that accurately describe the often convoluted binding site shapes
- Binding site protein residues that interact with ligands
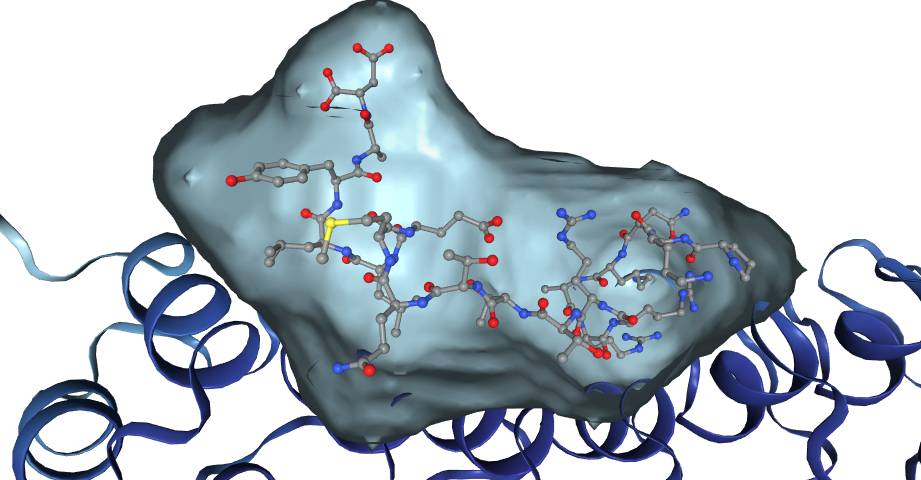
- Predicted ligands obtained using structure-based comparative ProBiS approach from similar binding sites in the PDB
- Binding site bounding box (in AutoDock Vina format) ready for docking
- Receptor, an AlphaFold2 predicted protein single chain structure
Input
- AlphaFold ID, UniProt ID, PDB ID and Chain ID (where available)
- Protein name
- Protein function, such as, protein kinases or cancer-related proteins
- Binding site type and binding site rank
- See tutorial for more query options